Recurrent patterns discovery with GrammarViz 2.0 and command line
1. Introduction
We discuss same as in GUI tutorial use-cases which show GrammarViz utility in variable length time series pattern discovery.
2. Dataset used
Two datasets are used in this demo:
2.1. Winding dataset
According to the original dataset source this dataset is a snapshot of data collected from industrial winding process whose column #2 corresponds to traction reel angular speed:
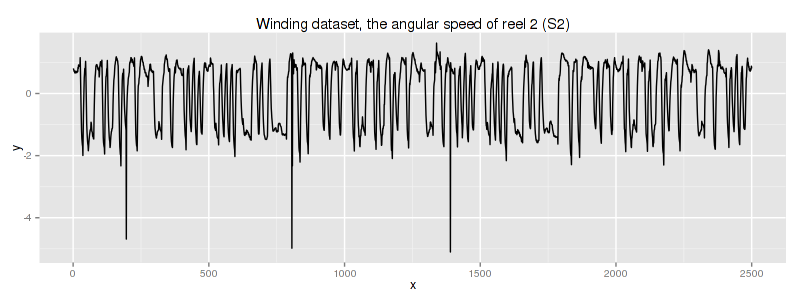
unfortunately, we do not know any specific information about this dataset.
2.2. QTDB 0606 ECG dataset
This data set (database record) can be downloaded from PHYSIONET FTP and converted into the text format by executing this command
rdsamp -r sele0606 -f 120.000 -l 60.000 -p -c | sed -n '701,3000p' >0606.csv
in the linux shell (assuming that you have rdsamp installed at your system). We use the second column of this file. This is our dataset overview:
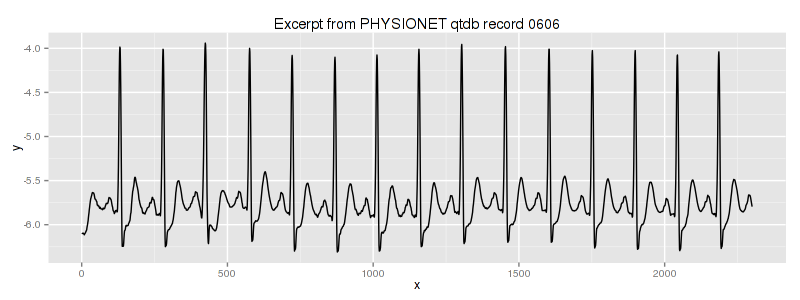
We know, that the third heartbeat of this ECG dataset contains the true anomaly as it was discussed in HOTSAX paper by Eamonn Keogh, Jessica Lin, and Ada Fu. Note, that the authors were specifically interested in finding anomalies which are shorter than a regular heartbeat following a suggestion given by the domain expert: “… We conferred with cardiologist, Dr. Helga Van Herle M.D., who informed us that heart irregularities can sometimes manifest themselves at scales significantly shorter than a single heartbeat….” Figure 13 of the paper further explains the nature of this true anomaly:
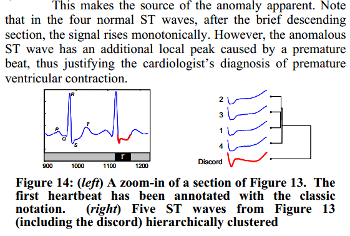
3. Using CLI.
By following the build instructions from our source code repository you can build a stand-alone jar file which we are going to use. It is named as grammarviz2-X.X.X-SNAPSHOT-jar-with-dependencies.jar where X.X.X is the latest release version. For this tutorial I have used the version tagged with
cli_pre-release tag which you can checkout from repo by running $ git checkout cli_pre-release.
Running the jar as usual, i.e., java -jar ... runs GUI, thus we must specify a specific class name (the one implementing the CLI interface). If run without parameters, this will print a short help message:
$ java -cp "target/grammarviz2-0.0.1-SNAPSHOT-jar-with-dependencies.jar" net.seninp.grammarviz.cli.TS2SequiturGrammar
Usage: <'main class'> [options]
Options:
--alphabet_size, -a
SAX alphabet size
Default: 4
--data_in, -d
The input file name
--data_out, -o
The output file name
--help, -h
Default: false
--strategy
Numerosity reduction strategy
Default: NONE
Possible Values: [NONE, EXACT, MINDIST]
--threshold
Normalization threshold
Default: 0.01
--window_size, -w
Sliding window size
Default: 30
--word_size, -p
PAA word size
Default: 6
4. Variable length recurrent patterns discovery
We use winding dataset in this tutorial with parameters of sliding window 100, PAA size 4 , alphabet size 3, and EXACT numerosity reduction strategy:
$ java -cp "target/grammarviz2-0.0.1-SNAPSHOT-jar-with-dependencies.jar" net.seninp.grammarviz.cli.TS2SequiturGrammar -d data/winding_col.txt -o winding_grammar.txt -w 100 -p 4 -a 3 --strategy EXACT GrammarViz2 CLI converter v.1 parameters: input file: data/winding_col.txt output file: winding_grammar.txt SAX sliding window size: 100 SAX PAA size: 4 SAX alphabet size: 3 SAX numerosity reduction: EXACT SAX normalization threshold: 0.01 14:19:28.906 [main] INFO n.s.g.cli.TS2SequiturGrammar - Reading data ... 14:19:29.102 [main] INFO n.s.g.cli.TS2SequiturGrammar - read 2500 points from null 14:19:29.102 [main] INFO n.s.g.cli.TS2SequiturGrammar - Performing SAX conversion ... 14:19:29.198 [main] INFO n.s.g.cli.TS2SequiturGrammar - Inferring Sequitur grammar ... 14:19:29.309 [main] INFO n.s.g.cli.TS2SequiturGrammar - Collecting stats ... 14:19:29.320 [main] INFO n.s.g.cli.TS2SequiturGrammar - Producing the output ...
This will yield a file named winding_grammar.txt containing the inferred grammar description and corresponding to its rules subsequences.
The rule #29 description from this file demonstrates the algorithm’s ability to capture repeated patterns of a length much larger than the original sliding window size. The subsequences corresponding to this rule are of length 354 and 353:
... /// R29 R29 -> 'R50 abbc abcc abcb R6 R46 R8 R13 R4 cacb R45 R52 R6 R16 bcba R20 R1 R2 acbb acba acbb', expanded rule string: 'bbac bbbc abbc abcc abcb bbcb bbca bcca bcba bcbb bcab ccab cbab cbac caac caab cbab cbbb cabb cacb bacb bbcb bbbb bbbc abbc acbc acbb abbb bbbb bbcb bbca cbca cbba bbba bcba ccaa cbaa cbab cbac caac baac babc babb bacb bbcb bbbb abbb acbb acba acbb ' subsequences starts: [806, 1806] subsequences lengths: [354, 353] rule occurrence frequency 2 rule use frequency 2 min length 353 max length 354 mean length 353 ...
at the same time, the subsequences corresponding to the most frequent rule #8 vary in length between 102 and 121 points:
... /// R8 R8 -> 'bcbb bcab', expanded rule string: 'bcbb bcab ' subsequences starts: [61, 115, 341, 565, 640, 852, 1565, 1640, 1853, 2060, 2341] subsequences lengths: [102, 103, 121, 109, 107, 114, 109, 108, 113, 103, 120] rule occurrence frequency 11 rule use frequency 5 min length 102 max length 121 mean length 109 ...
Similarly, if we use qtdb0606 dataset with SAX discretization parameters set to sliding window 100, PAA 8, and alphabet 4 (java -cp "target/grammarviz2-0.0.1-SNAPSHOT-jar-with-dependencies.jar" net.seninp.grammarviz.cli.TS2SequiturGrammar -d data/ecg0606_1.csv -o ecg_grammar.txt -w 100 -p 8 -a 4 --strategy EXACT), the algorithm finds that the most frequently occurring rules are normal heartbeats ranging in length from 105 to 107 points:
... /// R6 R6 -> 'bbcbbdab bbcbcdab', expanded rule string: 'bbcbbdab bbcbcdab ' subsequences starts: [62, 209, 506, 652, 798, 943, 1087, 1234, 1384, 1533, 1682, 1828, 1973, 2115] subsequences lengths: [105, 107, 107, 105, 107, 106, 106, 106, 106, 107, 106, 107, 107, 107] rule occurrence frequency 14 rule use frequency 4 min length 105 max length 107 mean length 106 ...
5. Discussion
Note, that due to two factors: the numerosity reduction embedded in the data discretization process and the nature of GI algorithms, that create rules based on the long-range correlations, the shown above recurrent pattern discovery technique yields sets of frequent subsequences of a variable length.